Request More Information
Oligonucleotide Analysis

While the world of biomedicines is largely made up of proteinaceous platforms, therapeutic oligonucleotides are becoming more prominent and more diverse each year. These classes of biomedicines are synthetic molecules with a chemical backbone structure composed of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) bases. These biomedicines can be designed to target specific mRNAs, miRNAs and proteins related to various diseases. While having the targeted specificity of antibody therapeutics, they can also be synthesized chemically like “low-molecular weight” drugs which reduces manufacturing costs compared to the biological process of protein-based biomedicines. Therapeutic oligonucleotides combine some of the best properties of “low-molecular weight” therapies and biopharmaceuticals.
The functions of nucleic acid medicines are various: small interfering RNA (siRNA) to control protein synthesis by coupling with mRNA, miRNA to strengthen miRNA functions, aptamers that bind to a protein to inhibit its functions, and ribozymes to directly cleave the target RNA are but a few. Their basic structure is a chain comprising a few dozen (deoxy)nucleotides including adenine, thymine, guanine, cytosine, and uracil, which are components of DNA and RNA.
Though these nucleic acid-based medicines can be chemically synthesized without the need to culture cells as in antibody medicines, they still have a range of complex molecular properties to be monitored. Purity, molecular weight, and sequence confirmation must be assessed before being used for in vitro or in vivo testing. These molecules are medium-sized having a molecular weight ranging from several thousands to tens of thousands. The confirmation whether or not synthesized nucleic acid medicines are arranged in the intended base sequence is a critical quality characteristic to ensure the action of pharmaceuticals. Because natural oligonucleotides are very susceptible enzymatic degradation in a biological host, these biomedicines are modified to improve in vivo stability, target specificity, and overall potency. Some of these modifications on Therapeutic oligonucleotides include chemically modification, and phosphorothioation (oxygen atom replaced with sulfur atom on the phosphate moiety) , and even conjugation with N-acetylgalactosamine (GalNac).
With the complexity brought on by their chemical modifications and synthesis, many analytical techniques are employed to monitor both the progress and final product. Shimadzu offers a wide array of analytical technologies and workflows to enable the synthetic chemist and analytical scientist to meet their challenges.
LabSolutions Insight Biologics
Software for Oligonucleotide Sequence Characterization
LabSolutions Insight Biologics is a dedicated software platform for oligonucleotide characterization using the LCMS-9030 or LCMS-9050 quadrupole time-of-flight type (Q-TOF) mass spectrometer. Main product and impurity identification includes several core editors for sequence, nucleotides, linkers, ribose and base modifications. Together with processing and integration for target modifications, Insight Biologics is a comprehensive workspace for data review, processing, and reporting.
Oligonucleotide Therapeutics Solution Guide
Oligonucleotide therapeutics are nucleic acid polymers generally comprised of a few to several dozen bases (including modified bases) linked together. They are produced by chemical synthesis and act directly on organisms without being translated into proteins. Oligonucleotide therapeutics are characterized by the ability to target specific diseases. Another advantage is that it takes less time than conventional methods to find new therapeutic candidates because oligonucleotides are easy to design and synthesize. However, it is a practical problem that oligonucleotide therapeutics are degraded and excreted rapidly after administration by exonucleases and endonucleases that are abundant in blood and cells. This problem is being addressed by the introduction of modified oligonucleotides to improve chemical stability in vivo and by the development of lesion-targeted DDS (drug delivery systems) technologies.
Featured Applications
Improving Oligonucleotide Analysis: Enhancing Mass Spectra Quality with LCMS-2050 Ion-Source Optimization
This application note presents a robust method for analyzing synthetic oligonucleotides, developed using optimized ion-source conditions on a Shimadzu LCMS2050 single quadrupole mass spectrometer to effectively overcome complex analytical challenges. The optimization of ion-source conditions is outlined, including adjustments to QarrayTM voltage, interface voltage, desolvation temperature, desolvation line temperature, and gas flow rates for nebulizing, drying and heating gasses. The optimization data demonstrate a notable reduction in common mobile phase adducts associated with ionpairing reverse-phase separation, while simultaneously increasing the total ion intensity of the non-adduct analyte signal.
Measurement of Oligonucleotide Impurities Using Shimadzu Nexera XS Inert Coupled To LCMS-9030 QTOF
The biopharmaceutical industry is experiencing significant growth in the market for oligonucleotide therapeutics, such as antisense oligonucleotides and small interfering RNA (siRNA). To effectively characterize these oligonucleotides, High-Resolution Mass Spectrometry (HRMS) plays a crucial role in ensuring their quality, safety, and efficacy. Typically, impurities in oligonucleotides can arise from several factors, which include incomplete synthesis, degradation, or chemical modifications during the manufacturing process. Therefore, regulatory guidelines mandate the analysis of oligonucleotide impurities to evaluate product quality and ensure its compliance with regulatory standards.
This application note presents an end-to-end workflow for quantifying oligonucleotide impurities by spiking the oligonucleotides into a Full-Length Product (FLP). To ensure complete inertness of the sample flow path for optimal chromatographic separation, Nexera XS inert UHPLC system (Fig. 1) is used along with bioinert-coating metal-free Shimpack ScepterTM Claris C18-300 column.
Oligonucleotide Characterization for Quality Control on the Shimadzu Single Quad LCMS-2050
Nucleic acid-based therapeutics have garnered increased attention in recent years as an innovative class of treatments in immunology, virology, and RNA-based therapies. It is crucial to differentiate between full-length and truncated nucleotides, as well as modified and unmodified versions for effective quality control. Traditional hybridization techniques, such as ELISA (enzyme-linked immunosorbent assay) and qPCR (polymerase chain reaction), offer sensitivity in detection, but lack specificity for effective impurity analysis. Liquid chromatography-mass spectrometry (LC-MS) is an advanced technique with inherent specificity that can be utilized to analyze impurities, degradants, and other biological/chemical modifications that cannot be analyzed by hybridization techniques. This application note outlines a workflow for confident mass confirmation across the entire range of nucleotide length oligomers (10-60mer), along with quantitation of yield using both UV and mass spectrometry data. The acquired data from these workflows were analyzed considering impurities, including aborted sequences (N-1, N-2) and mobile phase adducts.
Efficient Method Development of Oligonucleotides by Reversed-Phase Ion-Pair Chromatography
Nucleic acid drugs, such as antisense oligonucleotides, exert their effect by interacting with targets (genes and proteins) inside and outside of cells. Nucleic acid drugs are produced through chemical synthesis, but the synthesis process can introduce impurities such as shorter and longer length of products and protection groups. Therefore, proper separation of the target oligonucleotide is required. For LC separation, one commonly used mode is reversed-phase ion-pair chromatography (RP-IP). The separation patterns obtained with RP-IP chromatography can vary depending on the concentration of the ion pair reagent and the composition of organic solvent. In addition, the separation behavior can differ based on the length of products, nucleobase, and the presence of modifications. Therefore, it is important to optimize the separation for each sequence of oligonucleotides. This article describes how to achieve the optimal separation of oligonucleotides and related impurities efficiently by utilizing LabSolutionsMD, a dedicated software for supporting method development, through initial screening and optimization phase respectively.
Ion-Pair Reversed-Phase (IP-RP) LCMS-9030 (Q-TOF) Mass Spectrometer for Separation and Identification of Oligonucleotides
Oligonucleotide therapeutics are short synthetic DNA or RNA polymers with the potential to treat a wide range of diseases. Currently, 13 oligonucleotide therapeutics have been granted new drug approval (NDA) by the U.S. Food and Drug Administration (FDA). On December 11, 2020, the US. FDA issued the first emergency use authorization (EUA) for the Pfizer-BioNTech COVID-19 mRNA Vaccine to be used in the US, which has further spurred interest in the oligonucleotide therapeutics. Typical oligonucleotides therapeutics are approximately 15-30 bases, except aptamer oligonucleotides, which are commonly 40-60 bases. While shorter oligonucleotides (less than 20 bases) can be easily resolved by HPLC, the separation of longer sequences becomes progressively more challenging. This article introduces an ion-pair reversed-phase (IP-RP) LC-MS system for high-resolution separation and high-accuracy mass analysis of oligonucleotides. Shimadzu LCMS-9030 (Q-TOF) mass spectrometer is used for LC-MS analysis.
Determination of Molecular Mass and Quantification of Oligonucleotide Therapeutics Using Quadrupole Time-of-Flight Mass Spectrometer LCMS-9030
Oligonucleotide therapeutics are synthetic oligonucleotides that demonstrate their medical efficacy through binding to target genes or target proteins that may be responsible for a range of diseases. To date, eight types of oligonucleotide therapeutics have been approved, many of which have a length of approximately 20 bases. This article introduces an example of analysis using the Q-TOF mass spectrometer, LCMS-9030. As an oligonucleotide therapeutic, the 2’-MOE modified oligonucleotide having 20 bases was used. Accurate mass spectrometry determined the molecular mass of the therapeutic with an error of 3 mDa (0.05 ppm). When a calibration curve was prepared using the MRM mode on the LCMS-9030 mass spectrometer, linearity was observed within a range of 1 - 1000 ng/mL.
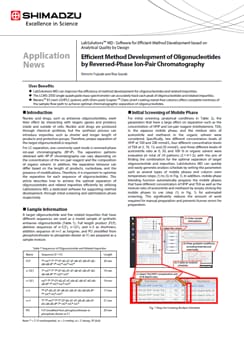
Quantitative Analysis and Determination of Molecular Weight of siRNA Type Oligonucleotides by LCMS™-8060
Nucleic acid drugs are synthetic oligonucleotides which are designed to bond specifically with target RNA or proteins. Although most nucleic acid drugs approved to date are the antisense type, the aptamer type and siRNA type have also been approved. Many nucleic acid drugs consist of around 20 bases and have molecular weights on the order of 6,000. Precision mass analysis techniques such as MALDI-TOF type and Q-TOF type LC/MS are used in measurements of the molecular weight of drug substances. On the other hand, the triple quadrupole mass spectrometer is generally used in quantitative analysis such as analysis of the fate of drugs in the blood, as this instrument offers high sensitivity in combination with a wide dynamic range. This article introduces an example of analysis of synthetic double-strand oligonucleotides using a Shimadzu LCMS- 8060 triple quadrupole mass spectrometer, assuming analysis of the drug substances of siRNA type nucleic acid drugs. Quantitativity was confirmed by SIM (selected ion monitoring). In preparation of the calibration curve by the SIM mode, linearity was confirmed in the range of 1 fmol to 10 pmol. In addition, the molecular weights were determined by deconvolution of a multivalent ion mass spectrum.
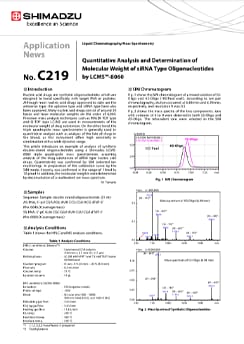
Verification of Quantitativity and Confirmation of Molecular Weight of Oligonucleotide Therapeutics Using LCMS™-8060
Oligonucleotide therapeutics are synthetic oligonucleotides which demonstrate medical efficacy by bonding with target genes or target proteins that cause various diseases. This article introduces an example of an analysis of a 20 base 2'-MOE modified oligonucleotide using an LCMS-8060 triple quadrupole mass spectrometer. In preparation of the calibration curve in the MRM mode, linearity was confirmed in the range of 1 to 300 ng/mL, and the molecular weight was confirmed by deconvolution from the multivalent ion mass spectrum obtained in the scan mode.
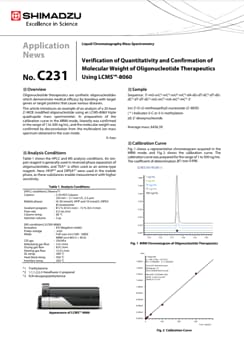
Analysis of Oligonucleotide Therapeutics using MALDI-8030 and LCMS-9030
Molecular characterization of nucleic acids using mass spectrometry is of current growing interest, because oligonucleotide therapeutics are a promising medicine to cure some diseases by working at upper stream of action mechanism with fewer side effects. Whereas a high-resolution accurate ESI mass spectrometer enables an exact intact mass measurement, routine oligo sequence analysis is a still hurdle, using ESI instruments. The complete internal oligo sequence is rarely obtained using typical ESI-MS/MS technique. On the other hand, in-source decay (ISD) using MALDI-TOFMS was reported as a useful method to conduct sequence analysis1) , although the instruments often employed do not have sufficient specification for exact mass measurement. Here, we report the characterization of oligonucleotide therapeutics using ESI-QTOF, LCMS-9030, and a dual polarity benchtop linear MALDI-TOFMS, MALDI-8030.
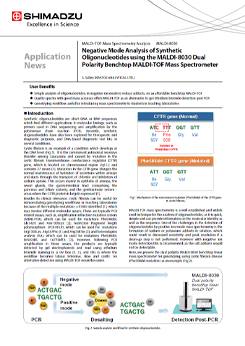
Negative Mode Analysis of Synthetic Oligonucleotides using the MALDI-8030 Dual Polarity Benchtop MALDI-TOF Mass Spectrometer
MALDI-TOF mass spectrometry is a well-established and widely used technique for the analysis of oligonucleotides, as it is quick, simple and can provide information on the molecular identity as well as the sequence. One of the challenges in the detection of oligonucleotides by positive ion mode mass spectrometry is the formation of sodium or potassium adducts in solution, which could result in decreased sensitivity and peak resolution if a clean-up step is not performed. However with negative ion mode detection this is circumvented, as the salt adducts would not be detectable. Here, we present the dual polarity MALDI-8030 benchtop linear mass spectrometer for genotyping, using cystic fibrosis disease (Phe508del mutation)as an example.
Synthesis Confirmation for Nucleic Acid Medicines - Rapid Sequence Confirmation Using a MALDI-TOF Mass Spectrometer
Medicines utilizing nucleic acids such as DNA and RNA that control genetic information are called "nucleic acid medicines". These nucleic acid medicines allow targeting of molecules such as messenger RNA (mRNA) and microRNA (miRNA) which cannot be targeted with traditional low-molecular-weight drugs and antibody medicines, and are expected to be innovative next generation pharmaceuticals for the treatment of genetic disorders which have been difficult to treat so far.
Analysis of Peptide Nucleic Acids (PNAs) using the MALDI-8020 benchtop linear MALDI-TOF mass spectrometer
Matrix assisted laser desorption/ionisation time-of- flight mass spectrometry (MALDI-TOF MS) is gaining popularity due to the ability of MALDI-MS to quickly provide information on accurate molecular mass using a simple sample preparation workflow. The MALDI-TOF MS can be used to confirm and guide synthesis of PNA oligomers, either in QC or R&D laboratories. Here, we demonstrate the analysis of three PNA samples using MALDI-8020 benchtop linear MALDI-TOF mass spectrometer.
Investigation of matrix conditions for nucleic acid analysis in positive ion detection using a linear benchtop MALDI-TOFMS
Nucleic acid is of interest in mass spectrometry, because it has been promising medicine to cure some diseases by working at upper stream of action mechanism with less side effect. Moreover, a molecular confirmation of nucleic acid using mass spectrometer is one of a routine analysis, where cost effectiveness of the instrument is significant factor. Generally negative ion detection is applied to the nucleic acid analysis. However, experimental conditions for positive ion detection have not been examined much, although recently a new bench-top instrument specialized to a linear mode with positive ion detection was introduced to achieve high cost-effectiveness. Here, the conditions for the positive ion detection will be reported using the benchtop mass spectrometer.
Oligonucleotides analysis by Ion Exchange Chromatography and Effects of pH Changes in the Mobile Phase on Separation
Nucleic acid is of interest in mass spectrometry, because it has been promising medicine to cure some diseases by working at upper stream of action mechanism with less side effect. Moreover, a molecular confirmation of nucleic acid using mass spectrometer is one of a routine analysis, where cost effectiveness of the instrument is significant factor. Generally negative ion detection is applied to the nucleic acid analysis. However, experimental conditions for positive ion detection have not been examined much, although recently a new bench-top instrument specialized to a linear mode with positive ion detection was introduced to achieve high cost-effectiveness. Here, the conditions for the positive ion detection will be reported using the benchtop mass spectrometer.
Thermal Stability Analysis of Nucleic Acid Drugs by TMSPC™-8 Tm Analysis System
Functional nucleic acids, beginning with nucleic acid drugs, have attracted considerable attention in recent years. In the development and evaluation of these functional nucleic acids, it is essential to understand their thermal stability, as this is a factor that controls their structure and functions. Thermal stability analysis is also indispensable in techniques based on hybridization of nucleic acids such as PCR and DNA microarray. In this article, a thermal stability analysis (Tm analysis) of a nucleic acid was conducted using a Shimadzu UV-1900i ultraviolet-visible (UV-Vis) spectrophotometer and TMSPC-8 Tm analysis system. Simple calculation of the Tm value of nucleic acids is possible by using this system.
Three-Dimensional Spectra Measurement of Fluorescent Probes used for DNA Detection
DNA probes labeled with fluorescent dye (below referred to as fluorescent probes) are used extensively to detect and identify specific DNA when conducting life science studies. The mechanism involves the selective binding of the probe to specific DNA, thereby permitting the detection of that DNA. However, due to the wide variety of fluorescent dyes, it is important to know the exact wavelength at which the probe fluoresces to ensure DNA detection. Here, using the three-dimensional spectral measurement feature of the RF-6000 Spectrofluorophotometer, we introduce examples of fluorescence measurement of two types of fluorescent probes.
Quantitation of Nucleic Acids – Trace Measurement Using TrayCell and Nano Stick –
Ultraviolet-visible (UV-VIS) spectrophotometers are used in quantitative and qualitative analysis of substances in many fields. Purity confirmation and quantitation of nucleic acids, proteins, and other substances are also performed in the life sciences, but measurement at the trace level is demanded, as only small samples are available in many cases. This article introduces an example of quantitation of nucleic acid with a Shimadzu UV-1900 UV-VIS spectrophotometer with two type of cells (TrayCellTM, Hellma Analytics and Nano Stick, SINCO) which enable measurement of sample quantities of several μL. The Biomethod Mode of the UV-1900 is also introduced briefly.
News / Events
-
AAPS Pharm Sci 360 2025
November 9-12
Henry B. Gonzalez Convention Center
San Antonio, TX -
ASMS (American Society for Mass Spectrometry) 2025
June 1-5
Baltimore Convention Center
Baltimore, MD -
Shimadzu Scientific Instruments Opens Boston Location of Its R&D Center Focus will be on promoting customer-oriented development to expand business in the pharmaceutical field
Shimadzu Scientific Instruments, Inc. (SSI, Columbia, Maryland, USA), a Shimadzu Group company, has opened a satellite lab in Boston, Massachusetts to be the base of its collaborative research and development activities on the East Coast. Established to conduct research and development more closely linked to customers, SSI's R&D Center consists of three bases, with the main facilities at its Maryland headquarters, a West Coast location, and this new space in Boston near the city center. The Boston lab was set up by partnering with Labshares, a shared laboratory service provider for life science companies.